Using the iNo score to discriminate normal from altered nucleolar morphology, with applications in basic cell biology and potential in human disease diagnostics
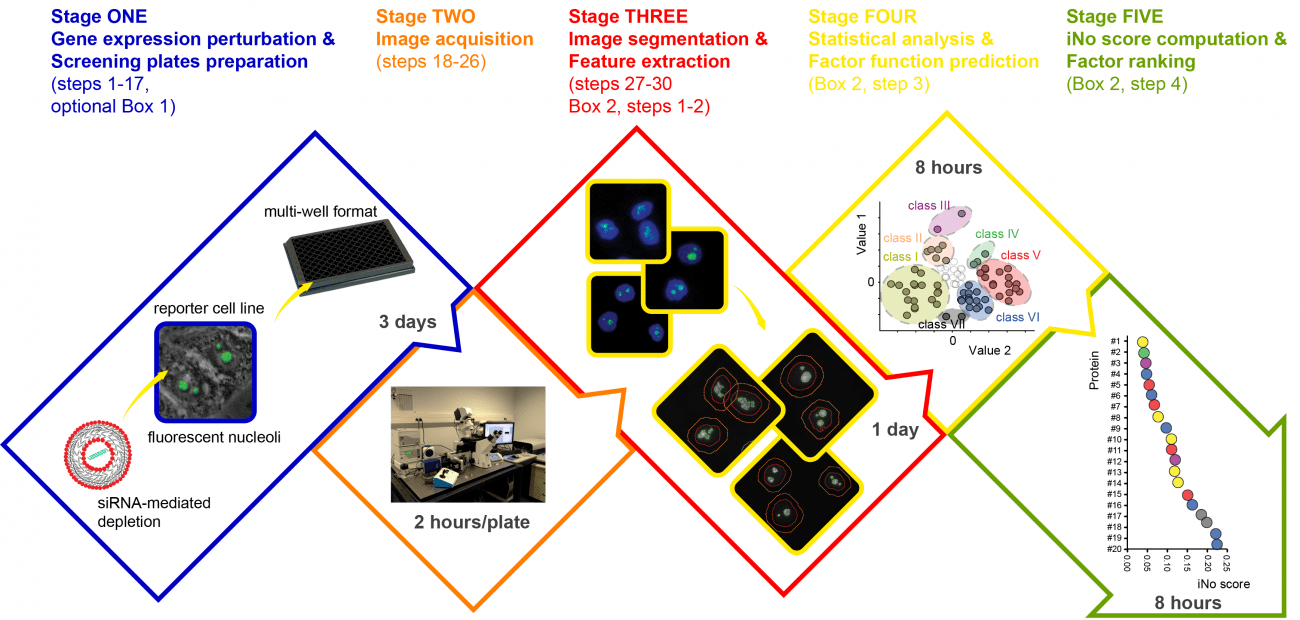
Ribosome biogenesis is initiated in the nucleolus, a cell condensate essential to gene expression, whose morphology informs cancer pathologists on the health status of a cell. Here we describe a protocol for assessing both qualitatively and quantitatively the involvement of trans-acting factors in nucleolar structure. The protocol involves depleting cells of factors of interest using small interfering RNAs, fluorescence imaging of nucleoli in an automated high-throughput platform, and use of a dedicated software to determine an index of nucleolar disruption, the iNo score. This scoring system is unique in that it integrates in a parametric equation the five most discriminant shape and textural features of the nucleolus. Determining the iNo score enables both qualitative and quantitative factor classification with prediction of function (functional clustering), which is not achieved by competing approaches, and stratification of their effect (severity of defects) on nucleolar structure. The iNo score has the potential to be useful in basic cell biology (nucleolar structure–function relationships, mitosis, and senescence), developmental and/or organismal biology (ageing), and clinical practice (cancer, viral infection, and reproduction). The entire protocol can be completed within 1 week.
See Stamatopoulou et al. (2018) Nature Protocols for details